This week, I spent some time to find out what NoWorkflow can actually do for us. So first, we start with a summary of NoWorkflow system including what kind of provenance can be captured and how we can access that. NoWorkflow system stores all the provenance captured in a local Sqlite database with the following tables (words in italic are table names, FK stands for foreign key):
Definition provenance (wf specification):
function_def: with schema (id, name, trial_id(FK to id in trial))
object: with schema (id, name, type, function_def_id(FK to id in function_def)); three types: argument, globe and function calls (internal functions)
Deployment provenance:
module: list of Python modules been used
dependency: showing which trial using which module
envorinment_attr: list of environmental attributes (JAVA_HOME, PID, etc.)
Execution provenance:
function_activation: same schema as in Prolog, FK to id in trial and FK to id in itself for caller_id indicating function call hierarchy.
object_value: with schema (id, name, value, type, function_activation_id(FK to id in function_activation)), two types: global and argument (indicating input data value to a function)
file_access: same schema as in Prolog, FK to id in trial and FK to id in function_activation indicating which function call triggered this file access
Other auxiliary tables:
trial: with trial id been referenced as foreign key
sqlite_sequence: not sure is useful information or not
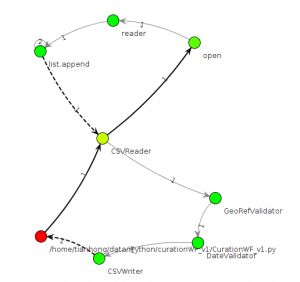
Of course we can directly query the Sqlite database to get the provenance information, but NoWorkflow system have several options that the provenance information can be exported into two different forms: Prolog rules and visualization graphs. Prolog rules have the same information of function_activation and file_access with the same schema but we can use logic rules to query the provenance and even some rules are provided by the NoWorkflow system. The visualization graph only show information of function activations that each node representing one function call, the edges indicating the control flow and the color of each node showing the relative runtime. An example as follows:
Another front is the use case from EVA WG. I’ve done with the translation from Matlab to Python, I will not say it’s a exact translation since some of the functions or configurations are slightly different between two languages, but the one I have will do 98% of the job. Here is an example output of the script in Python (dataset: BIOME-BGC_RG1_V1_Monthly_GPP_NA_2010.nc4):
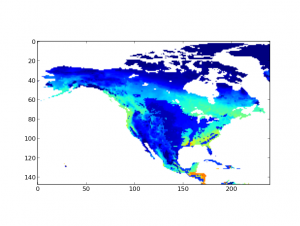
One big challenge here is that testing or running the script against some large dataset is time and space consuming. Unfortunately, most of the NetCDF data sets we have is really large since it captures the whole dataset of certain simulations which usually spans across multiple years on a large area.
Based on what we have on both of the above fronts, I think it’s a good time for the “big reunion” of these two fronts that we can now start to study the provenance of that particular use case. So the questions now is what kind of questions we can ask for this use case, how would provenance information helps us understand and improve this use case and of course how we achieve that.
As a start point, I have run NoWorkflow system on the Python script, but the result is actually not very existing. The reason is because of the following: 1) the script only read one file and no output file, so the file access provenance is just one “line”, and 2) the script itself is not “function oriented”, which means it’s one big function with a large “for” statement. So the function definition and function activation provenance is very straightforward. I think we should think about at which level or how fine-grained we’d like to capture provenance and we can define different functions for each portion of the code so NoWorkflow system can treat each portion of the code as different functions.
Though I wrote this Python script in iPython notebook but as I found several weeks ago, iPython notebook is not built to be a “heavy-duty” IDE and it’s difficult to split the script into different cells. So it’s one of the reasons why testing this script is little bit difficult and more importantly, there is not a lot of provenance information we can capture if we run the script in iPython notebook. But next week, I’ll start to look at two new tools we heard during Provenance Week 2014 last month. One tool is actually used to capture iPython notebook provenance, so that should be interesting and I’ll investigate how we can integrate this tool with ours.